Resource prioritization systems: Chicago food inspections#
Before continue, Did you…?
This case study, part of the dirtyduck tutorial, assumes that you already setup the tutorial’s infrastructure and load the dataset.
-
If you didn’t setup the infrastructure go here,
-
If you didn't load the data, you can do it very quickly or you can follow all the steps and explanations about the data.
Problem description#
We want to generate a list of facilities that will have a critical or serious food violation if inspected.
The scenario is the following: you work for the City of Chicago and you have limited food inspectors, so you try to prioritize them to focus on the highest-risk facilities. So you will use the data to answer the next question:
Which X facilities are most likely to fail a food inspection in the following Y period of time?
A more technical way of writing the question is: What is the probability distribution of facilities that are at risk of fail a food inspection if they are inspected in the following period of time?1
If you want to focus on major violations only, you can do that too:
Which X facilities are most likely to have a critical or serious violation in the following Y period of time?
This situation is very common in governmental agencies that provide social services: they need to prioritize their resources and use them in the facilities that are most likely to have problems
We will use machine learning to accomplish this. This means that we will use historical data to train our models, and we will use temporal cross validation to test the performance of them.
For the resource prioritization problems there are commonly two problems with the data: (a) bias and (b) incompleteness.
First, note that our data have bias: We only have data on facilities that were inspected. That means that our data set contains information about the probability of have a violation (V) given that the facility was inspected (I), P(V|I). But the probability that we try to find is P(V).
A different problem that our data set could have is if our dataset contains all the facilities in Chicago, i.e. if our entities table represents the Universe of facilities. There are almost 40,000 entities in our database. We could make the case that every facility in Chicago is in the database, since every facility that opens will be subject to an inspection. We will assume that all the facilities are in our data.
What do you want to predict?#
We are interested in two different outcomes:
- Which facilities are likely to fail an inspection?
The outcome takes a 1 if the inspection had at least one result = 'fail' and a 0 otherwise.
- Which facilities fail an inspection with a major violation?
Critical violations are coded between 1-14, serious violations
between 15-29, everything above 30 is assumed to be a minor
violation. The label takes a 1 if the inspection had at least one
result = 'fail' and a violation between 1 and 29, and a 0
otherwise.
I want to learn more about the data
Check the Data preparation section!
Data Changes
On 7/1/2018 the Chicago Department of Public Health’s Food Protection unit changed the definition of violations. The changes don’t affect structurally the dataset (e.g. how the violations are inputted to the database), but the redefinition will change the distribution and interpretation of the violation codes. See here.
We can extract the severity of the violation using the following code:
select
event_id,
entity_id,
date,
result,
array_agg(distinct obj ->>'severity') as violations_severity,
(result = 'fail') as failed,
coalesce(
(result = 'fail' and
('serious' = ANY(array_agg(obj ->> 'severity')) or 'critical' = ANY(array_agg(obj ->> 'severity')))
), false
) as failed_major_violation
from
(
select
event_id,
entity_id,
date,
result,
jsonb_array_elements(violations::jsonb) as obj
from
semantic.events
limit 20
) as t1
group by
entity_id, event_id, date, result
order by
date desc;
| event_id | entity_id | date | result | violations_severity | failed | failed_major_violation |
|---|---|---|---|---|---|---|
| 1770568 | 30841 | 2016-05-11 | pass | {minor} | f | f |
| 1763967 | 30841 | 2016-05-03 | fail | {critical,minor,serious} | t | t |
| 1434534 | 21337 | 2014-04-03 | pass | {NULL} | f | f |
| 1343315 | 22053 | 2013-06-06 | fail | {minor,serious} | t | t |
| 1235707 | 21337 | 2013-03-27 | pass | {NULL} | f | f |
| 537439 | 13458 | 2011-06-10 | fail | {NULL} | t | f |
| 569377 | 5570 | 2011-06-01 | pass | {NULL} | f | f |
The outcome will be used by triage to generate the labels
(once that we define a time span of interest). The
following image tries to show the meaning of the outcomes for the
inspection failed problem definition.
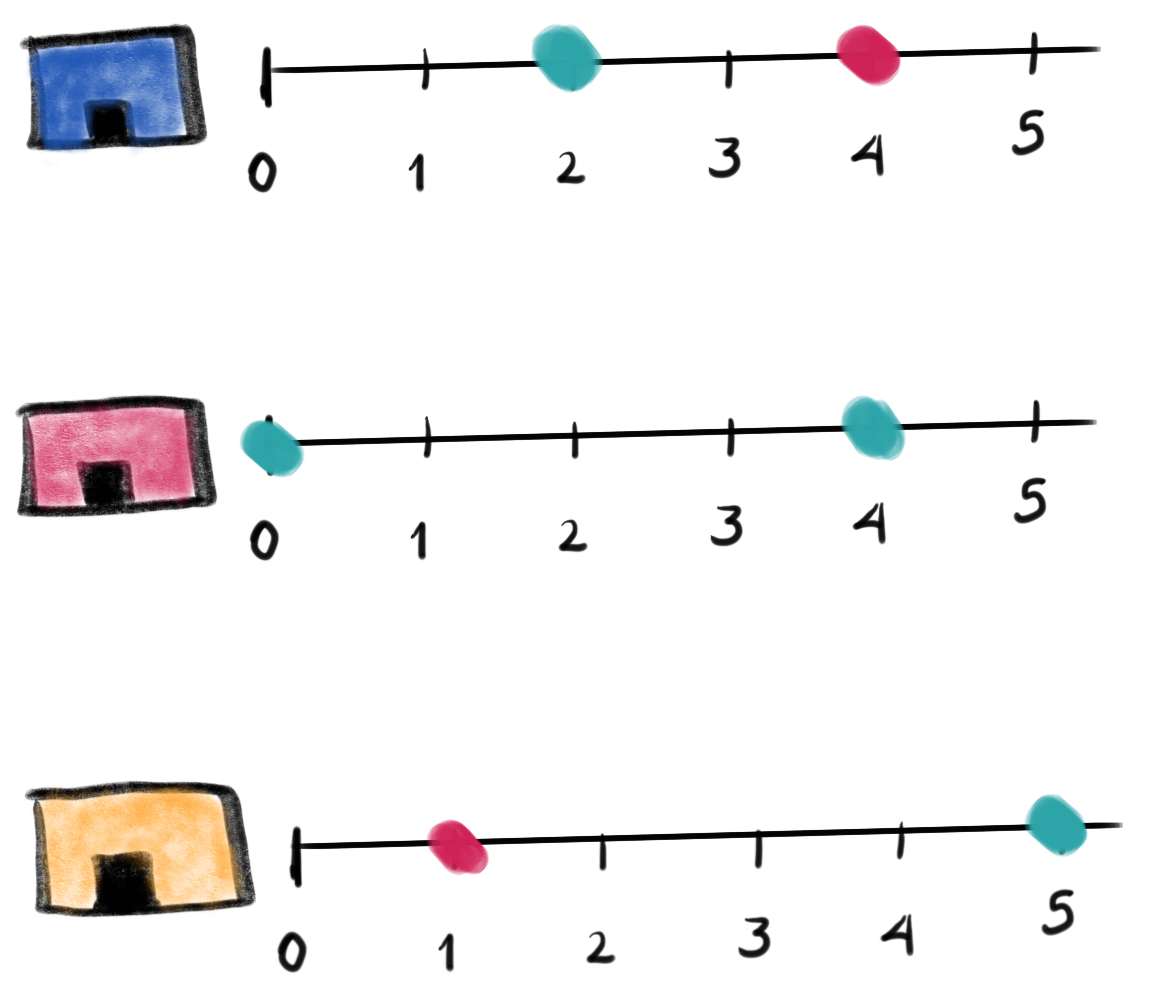 Figure. The image shows three
facilities (blue, red and orange) and, next to each, a temporal line with 6 days (0-5). Each
dot represents an inspection. Color is the **outcome* of the
inspection. Green means the facility passed the inspection, and red
means it failed. Each facility in the image had two inspections, but
only the facility in the middle passed both.*
Figure. The image shows three
facilities (blue, red and orange) and, next to each, a temporal line with 6 days (0-5). Each
dot represents an inspection. Color is the **outcome* of the
inspection. Green means the facility passed the inspection, and red
means it failed. Each facility in the image had two inspections, but
only the facility in the middle passed both.*
Modeling Using Machine Learning#
It is time to put these steps together. All the coding is complete
(triage dev team did that for us); we just need to modify the
triage experiment’s configuration file.
Defining a baseline#
As a first step, lets do an experiment that defines our
baseline. The rationale of this is that the knowing the baseline
will allow us to verify if our Machine Learning model is better than
the baseline. The baseline in our example will be a random selection
of facilities2. This is implemented as a DummyClassifier in
scikit-learn. Another advantage of starting with the baseline, is
that is very fast to train (DummyClassifier is
not computationally expensive) , so it will help us to verify that the
experiment configuration is correct without waiting for a long time.
Experiment description file
You could check the meaning about experiment description files (or configuration files) in A deeper look into triage.
We need to write the experiment config file for that. Let's break it down and explain their sections.
The config file for this first experiment is located in inspections_baseline.yaml.
The first lines of the experiment config file specify the config-file
version (v8 at the moment of writing this tutorial), a comment
(model_comment, which will end up as a value in the
triage_metadata.models table), and a list of user-defined metadata
(user_metadata) that can help to identify the resulting model
groups. For this example, if you run experiments that share a temporal
configuration but that use different label definitions (say, labeling
inspections with any violation as positive versus only labeling
inspections with major violations as positive), you can use the user
metadata keys to indicate that the matrices from these experiments
have different labeling criteria. The matrices from the two
experiments will have different filenames (and should not be
overwritten or incorrectly used), and if you add the
label_definition key to the model_group_keys, models made on
different label definitions will belong to different model groups.
config_version: 'v8'
model_comment: 'inspections: baseline'
random_seed: 23895478
user_metadata:
label_definition: 'failed'
file_name: 'inspections_baseline.yaml'
experiment_type: 'inspections prioritization'
description: |
Baseline calculation
purpose: 'baseline'
org: 'DSaPP'
team: 'Tutorial'
author: 'Your name here'
etl_date: '2019-05-07'
model_group_keys:
- 'class_path'
- 'parameters'
- 'feature_names'
- 'feature_groups'
- 'cohort_name'
- 'state'
- 'label_name'
- 'label_timespan'
- 'training_as_of_date_frequency'
- 'max_training_history'
- 'label_definition'
- 'experiment_type'
- 'org'
- 'team'
- 'author'
- 'etl_date'
Note
Obviously, change 'Your name here' for your name (if you like)
Next comes the temporal configuration section. The first four parameters are related to the availability of data: How much data you have for feature creation? How much data you have for label generation?
Data Changes
On 7/1/2018 the Chicago Department of Public Health’s Food Protection unit changed the definition of violations. The changes don’t affect structurally the dataset (e.g. how the violations are inputted to the database), but the redefinition will change the distribution and interpretation of the violation codes. See here.
The next parameters are related to the training intervals:
- How frequently to retrain models? (
model_update_frequency) - How many rows per entity in the train matrices? (
training_as_of_date_frequencies) - How much time is covered by labels in the training matrices? (
training_label_timespans)
The remaining elements are related to the testing matrices. For inspections, you can choose them as follows:
test_as_of_date_frequenciesis planning/scheduling frequencytest_durationshow far ahead do you schedule inspections?test_label_timespanis equal totest_durations
Let's assume that we need to do rounds of inspections every 6 months
(test_as_of_date_frequencies = 6month) and we need to complete that
round in exactly in that time (i.e. 6 months) (test_durations = test_label_timespan =
6month).
We will assume that the data is more or less stable3, at least for
now, so model_update_frequency = 6month.
temporal_config:
feature_start_time: '2010-01-04'
feature_end_time: '2018-06-01'
label_start_time: '2015-01-01'
label_end_time: '2018-06-01'
model_update_frequency: '6month'
training_label_timespans: ['6month']
training_as_of_date_frequencies: '6month'
test_durations: '0d'
test_label_timespans: ['6month']
test_as_of_date_frequencies: '6month'
max_training_histories: '5y'
We can visualize the time splitting using the function show-timechop
(See A deeper look into triage for more information)
# Remember to run this in bastion NOT in your laptop shell!
triage experiment experiments/inspections_baseline.yaml --show-timechop
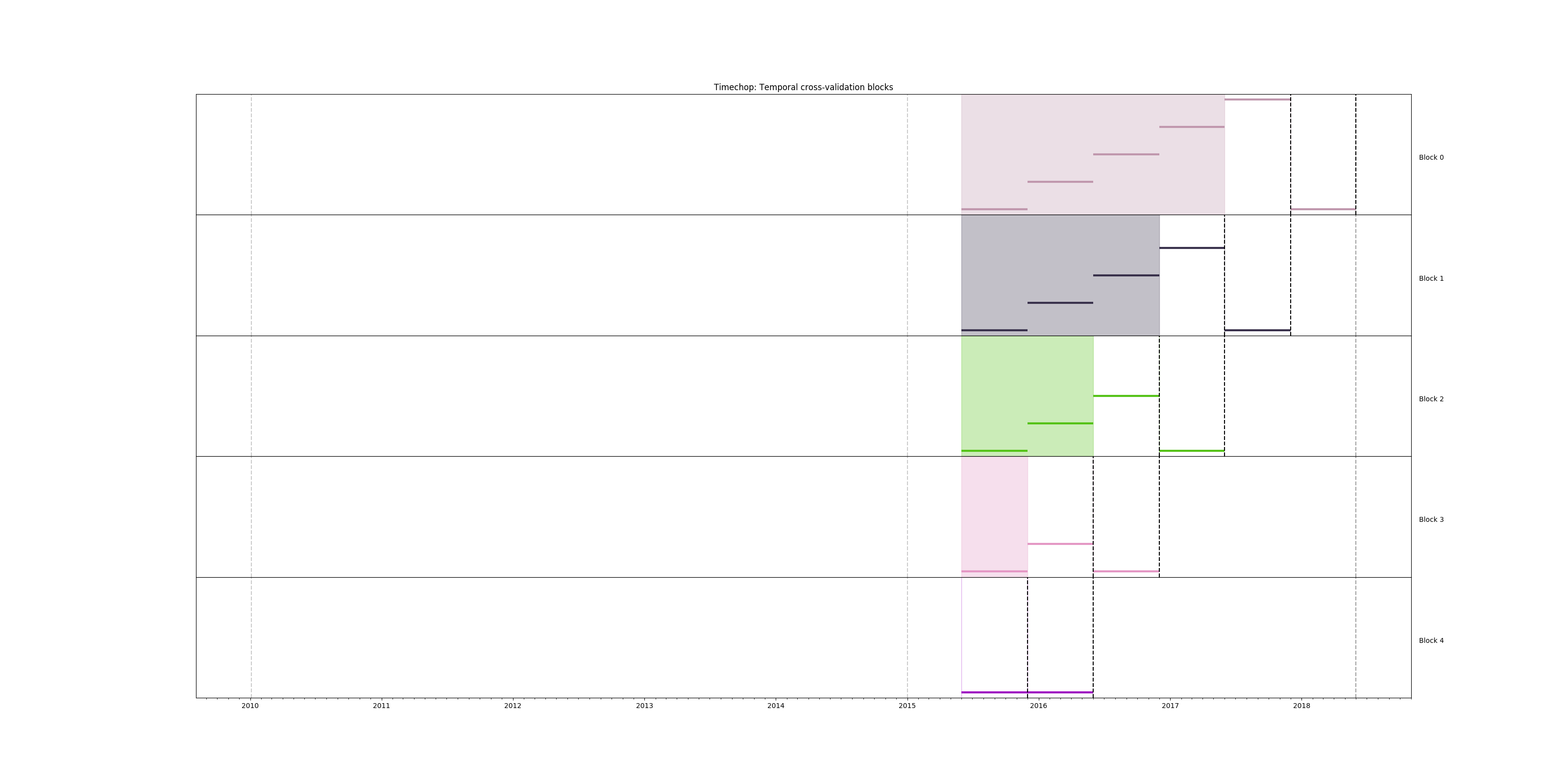 Figure. Temporal blocks for the inspections baseline experiment
Figure. Temporal blocks for the inspections baseline experiment
We need to specify our labels. For this first experiment we will use
the label failed, using the same query from the
simple_skeleton_experiment.yaml
label_config:
query: |
select
entity_id,
bool_or(result = 'fail')::integer as outcome
from semantic.events
where '{as_of_date}'::timestamp <= date
and date < '{as_of_date}'::timestamp + interval '{label_timespan}'
group by entity_id
name: 'failed_inspections'
It should be obvious, but let's state it anyway: We are only training
in facilities that were inspected, but we will test our model
in all the facilities in our cohort4. So, in the train matrices we
will have only 0 and 1 as possible labels, but in the test
matrices we will found 0, 1 and NULL.
I want to learn more about this…
In the section regarding to Early Warning Systems we will learn how to incorporate all the facilities of the cohort in the train matrices.
We just want to include active facilities in our matrices, so
we tell triage to take that in account:
cohort_config:
query: |
select e.entity_id
from semantic.entities as e
where
daterange(start_time, end_time, '[]') @> '{as_of_date}'::date
name: 'active_facilities'
Triage will generate the features for us, but we need to tell it
which features we want in the section feature_aggregations. Here,
each entry describes a collate.SpacetimeAggregation object and the
arguments needed to create it5. For this experiment, we will use only
one feature (number of inspections). DummyClassifier don't use any
feature to do the "prediction", so we won't expend compute cycles
doing the feature/matrix creation:
feature_aggregations:
-
prefix: 'inspections'
from_obj: 'semantic.events'
knowledge_date_column: 'date'
aggregates_imputation:
count:
type: 'zero_noflag'
aggregates:
-
quantity:
total: "*"
metrics:
- 'count'
intervals: ['all']
feature_group_definition:
prefix:
- 'inspections'
feature_group_strategies: ['all']
If we observe the image generated from the temporal_config section,
each particular date is the beginning of the rectangles that describes
the rows in the matrix. In that date (as_of_date in timechop
parlance) we will calculate the feature, and we will repeat that for
every other rectangle in that image.
Now, let's discuss how we will specify the models to try (remember
that the model is specified by the algorithm, the hyperparameters, and
the subset of features to use). In triage you need to specify in the
grid_config section a list of machine learning algorithms that you
want to train and a list of hyperparameters. You can use any algorithm
that you want; the only requirement is that it respects the sklearn
API.
grid_config:
'sklearn.dummy.DummyClassifier':
strategy: [uniform]
Finally, we should define wich metrics we care about for evaluating
our model. Here we will concentrate only in precision and recall
at an specific value k 6.
In this setting k represents the resource’s constraint: It is the number of inspections that the city could do in a month given all the inspectors available.
scoring:
testing_metric_groups:
-
metrics: [precision@, recall@, 'false negatives@', 'false positives@', 'true positives@', 'true negatives@']
thresholds:
percentiles: [1.0, 2.0, 3.0, 4.0, 5.0, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100]
top_n: [1, 5, 10, 25, 50, 100, 250, 500, 1000]
-
metrics: [auc, accuracy]
training_metric_groups:
-
metrics: [auc, accuracy]
-
metrics: [precision@, recall@]
thresholds:
percentiles: [1.0, 2.0, 3.0, 4.0, 5.0, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100]
top_n: [1, 5, 10, 25, 50, 100, 250, 500, 1000]
You should be warned that precision and recall at k in this
setting is kind of ill-defined (because you will end with a lot of
NULL labels, remember, only a few of facilities are inspected in
each period)7.
We will want a list of facilities to be inspected. The length of
our list is constrained by our inspection resources, i.e. the answer
to the question How many facilities can I inspect in a month? In
this experiment we are assuming that the maximum capacity is 10%
but we are evaluating for a larger space of possibilities (see
top_n, percentiles above).
The execution of the experiments can take a long time, so it is a good practice to validate the configuration file before running the model. You don't want to wait for hours (or days) and then discover that something went wrong.
# Remember to run this in bastion NOT in your laptop shell!
triage experiment experiments/inspections_baseline.yaml --validate-only
If everything was ok, you should see an Experiment validation ran to completion with no errors.
You can execute the experiment as8
# Remember to run this in bastion NOT in your laptop shell!
time triage experiment experiments/inspections_baseline.yaml
Protip
We are including the command time in order to get the total
running time of the experiment. You can remove it, if you like.
Don’t be scared!
This will print a lot of output! It is not an error!
We can query the table experiments to see the quantity of work that
triage needs to do
select
substring(experiment_hash, 1,4) as experiment,
config -> 'user_metadata' ->> 'description' as description,
total_features,
matrices_needed,
models_needed
from triage_metadata.experiments;
| experiment | description | total_features | matrices_needed | models_needed |
|---|---|---|---|---|
| e912 | "Baseline calculation\n" | 1 | 10 | 5 |
If everything is correct, triage will
create 10 matrices (5 for training, 5 for testing) in
triage/matrices and every matrix will be represented by two files,
one with the metadata of the matrix (a yaml file) and one with the
actual matrix (the gz file).
# We will use some bash magic
ls matrices | awk -F . '{print $NF}' | sort | uniq -c
Triage also will store 5 trained models in triage/trained_models:
ls trained_models | wc -l
And it will populate the results schema in the database. As
mentioned, we will get 1 model groups:
select
model_group_id,
model_type,
hyperparameters
from
triage_metadata.model_groups
where
model_config ->> 'experiment_type' ~ 'inspection'
| model_group_id | model_type | hyperparameters |
|---|---|---|
| 1 | sklearn.dummy.DummyClassifier | {"strategy": "prior"} |
And 5 models:
select
model_group_id,
array_agg(model_id) as models,
array_agg(train_end_time::date) as train_end_times
from
triage_metadata.models
where
model_comment ~ 'inspection'
group by
model_group_id
order by
model_group_id;
| model_group_id | models | train_end_times |
|---|---|---|
| 1 | {1,2,3,4,5} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} |
From that last query, you should note that the order in which triage
trains the models is from oldest to newest train_end_time and
model_group , also in ascending order. It will not go to the next
block until all the models groups are trained.
You can check on which matrix each models were trained:
select
model_group_id,
model_id, train_end_time::date,
substring(model_hash,1,5) as model_hash,
substring(train_matrix_uuid,1,5) as train_matrix_uuid,
ma.num_observations as observations,
ma.lookback_duration as feature_lookback_duration, ma.feature_start_time
from
triage_metadata.models as mo
join
triage_metadata.matrices as ma
on train_matrix_uuid = matrix_uuid
where
mo.model_comment ~ 'inspection'
order by
model_group_id,
train_end_time asc;
| model_group_id | model_id | train_end_time | model_hash | train_matrix_uuid | observations | feature_lookback_duration | feature_start_time |
|---|---|---|---|---|---|---|---|
| 1 | 1 | 2015-12-01 | 743a6 | 1c266 | 5790 | 5 years | 2010-01-04 00:00:00 |
| 1 | 2 | 2016-06-01 | 5b656 | 755c6 | 11502 | 5 years | 2010-01-04 00:00:00 |
| 1 | 3 | 2016-12-01 | 5c170 | 2aea8 | 17832 | 5 years | 2010-01-04 00:00:00 |
| 1 | 4 | 2017-06-01 | 55c3e | 3efdc | 23503 | 5 years | 2010-01-04 00:00:00 |
| 1 | 5 | 2017-12-01 | ac993 | a3762 | 29112 | 5 years | 2010-01-04 00:00:00 |
Each model was
trained with the matrix indicated in the column
train_matrix_uuid. This uuid is the file name of the stored
matrix. The model itself was stored under the file named with the
model_hash.
If you want to see in which matrix the model was tested you need to run the following query
select distinct
model_id,
model_group_id, train_end_time::date,
substring(model_hash,1,5) as model_hash,
substring(ev.matrix_uuid,1,5) as test_matrix_uuid,
ma.num_observations as observations
from
triage_metadata.models as mo
join
test_results.evaluations as ev using (model_id)
join
triage_metadata.matrices as ma on ev.matrix_uuid = ma.matrix_uuid
where
mo.model_comment ~ 'inspection'
order by
model_group_id, train_end_time asc;
| model_id | model_group_id | train_end_time | model_hash | test_matrix_uuid | observations |
|---|---|---|---|---|---|
| 1 | 1 | 2015-12-01 | 743a6 | 4a0ea | 18719 |
| 2 | 1 | 2016-06-01 | 5b656 | f908e | 19117 |
| 3 | 1 | 2016-12-01 | 5c170 | 00a88 | 19354 |
| 4 | 1 | 2017-06-01 | 55c3e | 8f3cf | 19796 |
| 5 | 1 | 2017-12-01 | ac993 | 417f0 | 20159 |
All the models were stored in /triage/trained_models/{model_hash}
using the standard serialization of sklearn models. Every model was
trained with the matrix train_matrix_uuid stored in the directory
/triage/matrices.
What's the performance of this model groups?
\set k 0.10 -- This defines a variable, "k = 0.10"
select distinct
model_group_id,
model_id,
ma.feature_start_time::date,
train_end_time::date,
ev.evaluation_start_time::date,
ev.evaluation_end_time::date,
to_char(ma.num_observations, '999,999') as observations,
to_char(ev.num_labeled_examples, '999,999') as "total labeled examples",
to_char(ev.num_positive_labels, '999,999') as "total positive labels",
to_char(ev.num_labeled_above_threshold, '999,999') as "labeled examples@k%",
to_char(:k * ma.num_observations, '999,999') as "predicted positive (PP)",
ARRAY[to_char(ev.best_value * ev.num_labeled_above_threshold,'999,999'),
to_char(ev.worst_value * ev.num_labeled_above_threshold,'999,999'),
to_char(ev.stochastic_value * ev.num_labeled_above_threshold, '999,999')]
as "true positive (TP)@k% (best,worst,stochastic)",
ARRAY[ to_char(ev.best_value, '0.999'), to_char(ev.worst_value, '0.999'),
to_char(ev.stochastic_value, '0.999')] as "precision@k% (best,worst,stochastic)",
to_char(ev.num_positive_labels*1.0 / ev.num_labeled_examples, '0.999') as baserate,
:k * 100 as "k%"
from
triage_metadata.models as mo
join
test_results.evaluations as ev using (model_id)
join
triage_metadata.matrices as ma on ev.matrix_uuid = ma.matrix_uuid
where
ev.metric || ev.parameter = 'precision@15_pct'
and
mo.model_comment ~ 'inspection'
order by
model_id, train_end_time asc;
order by
model_id, train_end_time asc;
| model_group_id | model_id | feature_start_time | train_end_time | evaluation_start_time | evaluation_end_time | observations | total labeled examples | total positive labels | labeled examples@k% | predicted positive (PP) | true positive (TP)@k% (best,worst,stochastic) | precision@k% (best,worst,stochastic) | baserate | k% |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 21 | 121 | 2010-01-04 | 2015-12-01 | 2015-12-01 | 2015-12-01 | 18,719 | 5,712 | 1,509 | 0 | 2,808 | {" 0"," 0"," 0"} | {" 0.000"," 0.000"," 0.000"} | 0.264 | 15.00 |
| 21 | 122 | 2010-01-04 | 2016-06-01 | 2016-06-01 | 2016-06-01 | 19,117 | 6,330 | 1,742 | 0 | 2,868 | {" 0"," 0"," 0"} | {" 0.000"," 0.000"," 0.000"} | 0.275 | 15.00 |
| 21 | 123 | 2010-01-04 | 2016-12-01 | 2016-12-01 | 2016-12-01 | 19,354 | 5,671 | 1,494 | 0 | 2,903 | {" 0"," 0"," 0"} | {" 0.000"," 0.000"," 0.000"} | 0.263 | 15.00 |
| 21 | 124 | 2010-01-04 | 2017-06-01 | 2017-06-01 | 2017-06-01 | 19,796 | 5,609 | 1,474 | 0 | 2,969 | {" 0"," 0"," 0"} | {" 0.000"," 0.000"," 0.000"} | 0.263 | 15.00 |
| 21 | 125 | 2010-01-04 | 2017-12-01 | 2017-12-01 | 2017-12-01 | 20,159 | 4,729 | 1,260 | 0 | 3,024 | {" 0"," 0"," 0"} | {" 0.000"," 0.000"," 0.000"} | 0.266 | 15.00 |
The columns num_labeled_examples, num_labeled_above_threshold,
num_positive_labels represent the number of selected entities on the
prediction date that are labeled, the number of entities with a
positive label above the threshold, and the number of entities with
positive labels among all the labeled entities respectively.
We added some extra columns: baserate, predicted positive (PP)
and true positive (TP). Baserate represents the proportion of the
all the facilities that were inspected that failed the inspection,
i.e. P(V|I). The PP and TP are approximate since it were
calculated using the value of k or the precision value. But you could get the exact value
of those from the test_results.predictions table.
Also note that in the precision@k% column we are showing three
numbers: best, worst, stochastic.
They try to answer the question How do you break ties in the
prediction score? This is important because it will affect the
calculation of your metrics. The Triage proposed solution to this is calculate the metric in the best case
scenario (score descending, all the true labels are at the top), and then do it in the worst
case scenario (score descending, all the true labels are at the
bottom) and then
calculate the metric several times (n=30) with the labels randomly shuffled
(a.k.a. stochastic scenario), so you
get the mean metric, plus some confidence intervals.
This problem is not specific of an inspection problem, is more related to
simple models like a shallow Decision Tree or a Dummy Classifier
when score ties likely will occur.
Note how in this model, the stochastic value is close to the baserate, since we are selecting at random using the prior.
Check this!
Note that the baserate should be equal to the precision@100%, if is not there is something wrong …
Creating a simple experiment#
We will try two of the simplest machine learning algorithms: a Decision Tree Classifier (DT) and a Scaled Logistic Regression (SLR)12 as a second experiment. The rationale of this is that the DT is very fast to train (so it will help us to verify that the experiment configuration is correct without waiting for a long time) and it helps you to understand the structure of your data.
The config file for this first experiment is located in /triage/experiments/inspections_dt.yaml
Note that we don't modify the temporal_config section neither the
feature_aggregations, cohort_config or label_config. Triage is
smart enough to use the previous tables and matrices instead of
generating them from scratch.
config_version: 'v8'
model_comment: 'inspections: basic ML'
user_metadata:
label_definition: 'failed'
experiment_type: 'inspections prioritization'
file_name: 'inspections_dt.yaml'
description: |
DT and SLR
purpose: 'data mining'
org: 'DSaPP'
team: 'Tutorial'
author: 'Your name here'
etl_date: '2019-02-21'
Note that we don't modify the temporal_config section neither the
cohort_config or label_config. Triage is smart enough to use the
previous tables and matrices instead of generating them from scratch.
For this experiment, we will add the following features:
-
Number of different types of inspections the facility had in the last year (calculated for an as-of-date).
-
Number of different types of inspections that happened in the zip code in the last year from a particular day.
-
Number of inspections
-
Number/proportion of inspections by result type
-
Number/proportion of times that a facility was classify with particular risk level
In all of them we will do the aggregation in the last month, 3 months, 6 months, 1 year and historically. Remember that all this refers to events in the past, i.e. How many times the facility was marked with high risk in the previous 3 Months?, What is the proportion of failed inspections in the previous year?
feature_aggregations:
-
prefix: 'inspections'
from_obj: 'semantic.events'
knowledge_date_column: 'date'
aggregates_imputation:
count:
type: 'zero_noflag'
aggregates:
-
quantity:
total: "*"
metrics:
- 'count'
intervals: ['1month', '3month', '6month', '1y', 'all']
-
prefix: 'risks'
from_obj: 'semantic.events'
knowledge_date_column: 'date'
categoricals_imputation:
sum:
type: 'zero'
avg:
type: 'zero'
categoricals:
-
column: 'risk'
choices: ['low', 'medium', 'high']
metrics:
- 'sum'
- 'avg'
intervals: ['1month', '3month', '6month', '1y', 'all']
-
prefix: 'results'
from_obj: 'semantic.events'
knowledge_date_column: 'date'
categoricals_imputation:
all:
type: 'zero'
categoricals:
-
column: 'result'
choice_query: 'select distinct result from semantic.events'
metrics:
- 'sum'
- 'avg'
intervals: ['1month', '3month', '6month', '1y', 'all']
-
prefix: 'inspection_types'
from_obj: 'semantic.events'
knowledge_date_column: 'date'
categoricals_imputation:
sum:
type: 'zero_noflag'
categoricals:
-
column: 'type'
choice_query: 'select distinct type from semantic.events where type is not null'
metrics:
- 'sum'
intervals: ['1month', '3month', '6month', '1y', 'all']
And as stated, we will train some Decision Trees, in particular we are interested in some shallow trees, and in a full grown tree. These trees will show you the structure of your data. We also will train some Scaled Logistic Regression, this will show us how "linear" is the data (or how the assumptions of the Logistic Regression holds in this data)
grid_config:
'sklearn.tree.DecisionTreeClassifier':
criterion: ['gini']
max_features: ['sqrt']
max_depth: [1, 2, 5,~]
min_samples_split: [2,10,50]
'triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression':
penalty: ['l1','l2']
C: [0.000001, 0.0001, 0.01, 1.0]
About yaml and sklearn
Some of the parameters in sklearn are None. If you want to try
those you need to indicate it with yaml's null or ~ keyword.
Besides the algorithm and the hyperparameters, you should specify
which subset of features use. First, in the section
feature_group_definition you specify how to group the features (you
can use the table name or the prefix from the section
feature_aggregation) and then a strategy for choosing the subsets:
all (all the subsets at once), leave-one-out (try all the subsets
except one, do that for all the combinations), or leave-one-in (just
try subset at the time).
feature_group_definition:
prefix:
- 'inspections'
- 'results'
- 'risks'
- 'inspection_types'
feature_group_strategies: ['all']
Finally we will leave the scoring section as before.
In this experiment we will end with 6 model groups (number of algorithms [1] \times number of hyperparameter combinations [2 \times 3 = 5] \times number of feature groups strategies [1]]). Also, we will create 18 models (3 per model group) given that we have 3 temporal blocks (one model per temporal group).
Before running the experiment, remember to validate that the configuration is correct:
# Remember to run this in bastion NOT in your laptop shell!
triage experiment experiments/inspections_dt.yaml --validate-only
and check the temporal cross validation:
# Remember to run this in bastion NOT in your laptop shell!
triage experiment experiments/inspections_dt.yaml --show-timechop
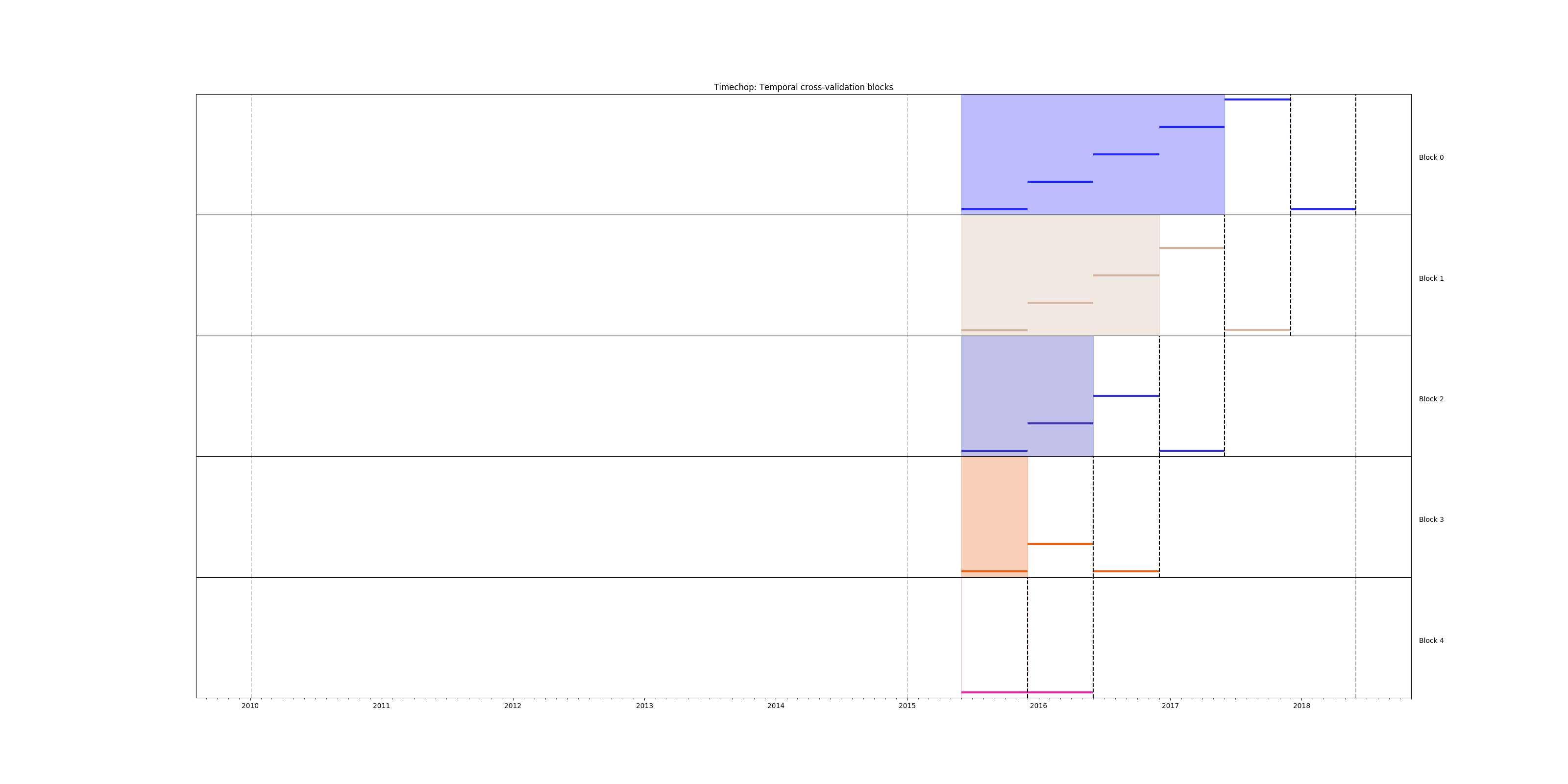 Temporal blocks for inspections experiment. The label is a failed inspection in the next year.
Temporal blocks for inspections experiment. The label is a failed inspection in the next year.
You can execute the experiment like this:
# Remember to run this in bastion NOT in your laptop shell!
time triage experiment experiments/inspections_dt.yaml
Again, we can run the following sql to see which things triage
needs to run:
select
substring(experiment_hash, 1,4) as experiment,
config -> 'user_metadata' ->> 'description' as description,
total_features,
matrices_needed,
models_needed
from triage_metadata.experiments;
| experiment | description | total_features | matrices_needed | models_needed |
|---|---|---|---|---|
| e912 | Baseline calculation | 1 | 10 | 5 |
| b535 | DT and SLR | 201 | 10 | 100 |
You can compare our two experiments and there are several differences, mainly in the order of magnitude. Like the number of features (1 vs 201) and models built (5 vs 100).
The After the experiment finishes, you will get 19 new model_groups (1 per combination in grid_config)
select
model_group_id,
model_type,
hyperparameters
from
triage_metadata.model_groups
where
model_group_id not in (1);
| model_group_id | model_type | hyperparameters |
|---|---|---|
| 2 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 1, "max_features": "sqrt", "min_samples_split": 2} |
| 3 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 1, "max_features": "sqrt", "min_samples_split": 10} |
| 4 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 1, "max_features": "sqrt", "min_samples_split": 50} |
| 5 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 2, "max_features": "sqrt", "min_samples_split": 2} |
| 6 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 2, "max_features": "sqrt", "min_samples_split": 10} |
| 7 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 2, "max_features": "sqrt", "min_samples_split": 50} |
| 8 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 5, "max_features": "sqrt", "min_samples_split": 2} |
| 9 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 5, "max_features": "sqrt", "min_samples_split": 10} |
| 10 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": 5, "max_features": "sqrt", "min_samples_split": 50} |
| 11 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": null, "max_features": "sqrt", "min_samples_split": 2} |
| 12 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": null, "max_features": "sqrt", "min_samples_split": 10} |
| 13 | sklearn.tree.DecisionTreeClassifier | {"criterion": "gini", "max_depth": null, "max_features": "sqrt", "min_samples_split": 50} |
| 14 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 0.000001, "penalty": "l1"} |
| 15 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 0.000001, "penalty": "l2"} |
| 16 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 0.0001, "penalty": "l1"} |
| 17 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 0.0001, "penalty": "l2"} |
| 18 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 0.01, "penalty": "l1"} |
| 19 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 0.01, "penalty": "l2"} |
| 20 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 1.0, "penalty": "l1"} |
| 21 | triage.component.catwalk.estimators.classifiers.ScaledLogisticRegression | {"C": 1.0, "penalty": "l2"} |
and 100 models (as stated before)
select
model_group_id,
array_agg(model_id) as models,
array_agg(train_end_time) as train_end_times
from
triage_metadata.models
where
model_group_id not in (1)
group by
model_group_id
order by
model_group_id;
| model_group_id | models | train_end_times |
|---|---|---|
| 2 | {6,26,46,66,86} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 3 | {7,27,47,67,87} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 4 | {8,28,48,68,88} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 5 | {9,29,49,69,89} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 6 | {10,30,50,70,90} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 7 | {11,31,51,71,91} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 8 | {12,32,52,72,92} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 9 | {13,33,53,73,93} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 10 | {14,34,54,74,94} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 11 | {15,35,55,75,95} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 12 | {16,36,56,76,96} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 13 | {17,37,57,77,97} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 14 | {18,38,58,78,98} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 15 | {19,39,59,79,99} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 16 | {20,40,60,80,100} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 17 | {21,41,61,81,101} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 18 | {22,42,62,82,102} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 19 | {23,43,63,83,103} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 20 | {24,44,64,84,104} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
| 21 | {25,45,65,85,105} | {"2015-12-01 00:00:00","2016-06-01 00:00:00","2016-12-01 00:00:00","2017-06-01 00:00:00","2017-12-01 00:00:00"} |
Let's see the performance over time of the models so far:
select
model_group_id,
array_agg(model_id order by ev.evaluation_start_time asc) as models,
array_agg(ev.evaluation_start_time::date order by ev.evaluation_start_time asc) as evaluation_start_time,
array_agg(ev.evaluation_end_time::date order by ev.evaluation_start_time asc) as evaluation_end_time,
array_agg(to_char(ev.num_labeled_examples, '999,999') order by ev.evaluation_start_time asc) as labeled_examples,
array_agg(to_char(ev.num_labeled_above_threshold, '999,999') order by ev.evaluation_start_time asc) as labeled_above_threshold,
array_agg(to_char(ev.num_positive_labels, '999,999') order by ev.evaluation_start_time asc) as total_positive_labels,
array_agg(to_char(ev.stochastic_value, '0.999') order by ev.evaluation_start_time asc) as "precision@15%"
from
triage_metadata.models as mo
inner join
triage_metadata.model_groups as mg using(model_group_id)
inner join
test_results.evaluations as ev using(model_id)
where
mg.model_config ->> 'experiment_type' ~ 'inspection'
and
ev.metric||ev.parameter = 'precision@15_pct'
and model_group_id between 2 and 21
group by
model_group_id
| model_group_id | models | evaluation_start_time | evaluation_end_time | labeled_examples | labeled_above_threshold | total_positive_labels | precision@15% |
|---|---|---|---|---|---|---|---|
| 1 | {1,2,3,4,5} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 0"," 0"," 0"," 0"," 0"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.000"," 0.000"," 0.000"," 0.000"," 0.000"} |
| 2 | {6,26,46,66,86} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 0"," 578"," 730"," 574"," 0"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.000"," 0.352"," 0.316"," 0.315"," 0.000"} |
| 3 | {7,27,47,67,87} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 161"," 622"," 0"," 433"," 0"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.283"," 0.203"," 0.000"," 0.282"," 0.000"} |
| 4 | {8,28,48,68,88} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 161"," 1,171"," 0"," 0"," 995"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.285"," 0.348"," 0.000"," 0.000"," 0.306"} |
| 5 | {9,29,49,69,89} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 726"," 1,318"," 362"," 489"," 291"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.306"," 0.360"," 0.213"," 0.294"," 0.241"} |
| 6 | {10,30,50,70,90} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 992"," 730"," 0"," 176"," 290"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.294"," 0.349"," 0.000"," 0.324"," 0.334"} |
| 7 | {11,31,51,71,91} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 1,023"," 325"," 329"," 176"," 1,033"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.323"," 0.400"," 0.347"," 0.323"," 0.315"} |
| 8 | {12,32,52,72,92} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 1,250"," 1,013"," 737"," 1,104"," 939"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.331"," 0.280"," 0.327"," 0.335"," 0.365"} |
| 9 | {13,33,53,73,93} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 595"," 649"," 547"," 1,000"," 841"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.281"," 0.250"," 0.309"," 0.350"," 0.356"} |
| 10 | {14,34,54,74,94} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 1,012"," 887"," 856"," 387"," 851"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.345"," 0.339"," 0.342"," 0.248"," 0.327"} |
| 11 | {15,35,55,75,95} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 0"," 0"," 0"," 0"," 0"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.000"," 0.000"," 0.000"," 0.000"," 0.000"} |
| 12 | {16,36,56,76,96} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 1,094"," 1,033"," 878"," 880"," 842"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.278"," 0.332"," 0.285"," 0.311"," 0.299"} |
| 13 | {17,37,57,77,97} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 995"," 1,117"," 987"," 623"," 651"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.313"," 0.324"," 0.320"," 0.318"," 0.299"} |
| 14 | {18,38,58,78,98} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 0"," 0"," 0"," 0"," 0"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.000"," 0.000"," 0.000"," 0.000"," 0.000"} |
| 15 | {19,39,59,79,99} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 771"," 582"," 845"," 570"," 625"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.246"," 0.227"," 0.243"," 0.244"," 0.232"} |
| 16 | {20,40,60,80,100} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 0"," 0"," 0"," 0"," 0"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.000"," 0.000"," 0.000"," 0.000"," 0.000"} |
| 17 | {21,41,61,81,101} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 783"," 587"," 813"," 586"," 570"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.250"," 0.235"," 0.253"," 0.259"," 0.253"} |
| 18 | {22,42,62,82,102} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 551"," 649"," 588"," 552"," 444"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.310"," 0.336"," 0.355"," 0.330"," 0.372"} |
| 19 | {23,43,63,83,103} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 1,007"," 776"," 818"," 784"," 725"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.343"," 0.409"," 0.373"," 0.366"," 0.421"} |
| 20 | {24,44,64,84,104} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 797"," 971"," 887"," 770"," 745"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.311"," 0.355"," 0.347"," 0.395"," 0.431"} |
| 21 | {25,45,65,85,105} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {2015-12-01,2016-06-01,2016-12-01,2017-06-01,2017-12-01} | {" 5,712"," 6,330"," 5,671"," 5,609"," 4,729"} | {" 943"," 953"," 837"," 730"," 699"} | {" 1,509"," 1,742"," 1,494"," 1,474"," 1,260"} | {" 0.320"," 0.363"," 0.349"," 0.397"," 0.429"} |
Which model in production (model selection) is something that we
will review later, with Audition, but for now, let's choose the
model group 3 and see the predictions table:
select
model_id,
entity_id,
as_of_date::date,
round(score,2),
label_value as label
from
test_results.predictions
where
model_id = 11
order by as_of_date asc, score desc
limit 20
| model_id | entity_id | as_of_date | round | label |
|---|---|---|---|---|
| 11 | 26873 | 2015-06-01 | 0.49 | |
| 11 | 26186 | 2015-06-01 | 0.49 | |
| 11 | 25885 | 2015-06-01 | 0.49 | |
| 11 | 24831 | 2015-06-01 | 0.49 | |
| 11 | 24688 | 2015-06-01 | 0.49 | |
| 11 | 21485 | 2015-06-01 | 0.49 | |
| 11 | 20644 | 2015-06-01 | 0.49 | |
| 11 | 20528 | 2015-06-01 | 0.49 | |
| 11 | 19531 | 2015-06-01 | 0.49 | |
| 11 | 18279 | 2015-06-01 | 0.49 | |
| 11 | 17853 | 2015-06-01 | 0.49 | |
| 11 | 17642 | 2015-06-01 | 0.49 | |
| 11 | 16360 | 2015-06-01 | 0.49 | |
| 11 | 15899 | 2015-06-01 | 0.49 | |
| 11 | 15764 | 2015-06-01 | 0.49 | |
| 11 | 15381 | 2015-06-01 | 0.49 | |
| 11 | 15303 | 2015-06-01 | 0.49 | |
| 11 | 14296 | 2015-06-01 | 0.49 | |
| 11 | 14016 | 2015-06-01 | 0.49 | |
| 11 | 27627 | 2015-06-01 | 0.49 |
NOTE: Given that this is a shallow tree, there will be a lot of
entities with the same score,you probably will get a different set
of entities, since postgresql will sort them at random.
It is important to know…
Triage sorted the predictions at random using the
random_seed from the experiment’s config file. If you want the
predictions being sorted in a different way add
prediction:
randk_tiebreaker: "worst" # or "best" or "random"
Note that at the top of the list (sorted by as_of_date, and then by
score), the labels are NULL. This means that the facilities that
you are classifying as high risk, actually weren't inspected in that
as of date. So, you actually don't know if this is a correct
prediction or not.
This is a characteristic of all the resource optimization problems: You do not have all the information about the elements in your system9.
So, how the precision/recall is calculated? The number that is show in
the evaluations table is calculated using only the rows that have a
non-null label. You could argue that this is fine, if you assume that
the distribution of the label in the non-observed facilities is the
same that the ones that were inspected that month10. We will come
back to this problem in the Early Warning Systems.
A more advanced experiment#
Ok, let's add a more complete experiment. First the usual generalities.
config_version: 'v8'
model_comment: 'inspections: advanced'
user_metadata:
label_definition: 'failed'
experiment_type: 'inspections prioritization'
description: |
Using Ensamble methods
purpose: 'trying ensamble algorithms'
org: 'DSaPP'
team: 'Tutorial'
author: 'Your name here'
etl_date: '2019-02-21'
We won't change anything related to features, cohort and label definition neither to temporal configuration.
As before, we can check the temporal structure of our crossvalidation:
# Remember to run this in bastion NOT in your laptop shell!
triage experiment experiments/inspections_label_failed_01.yaml --show-timechop
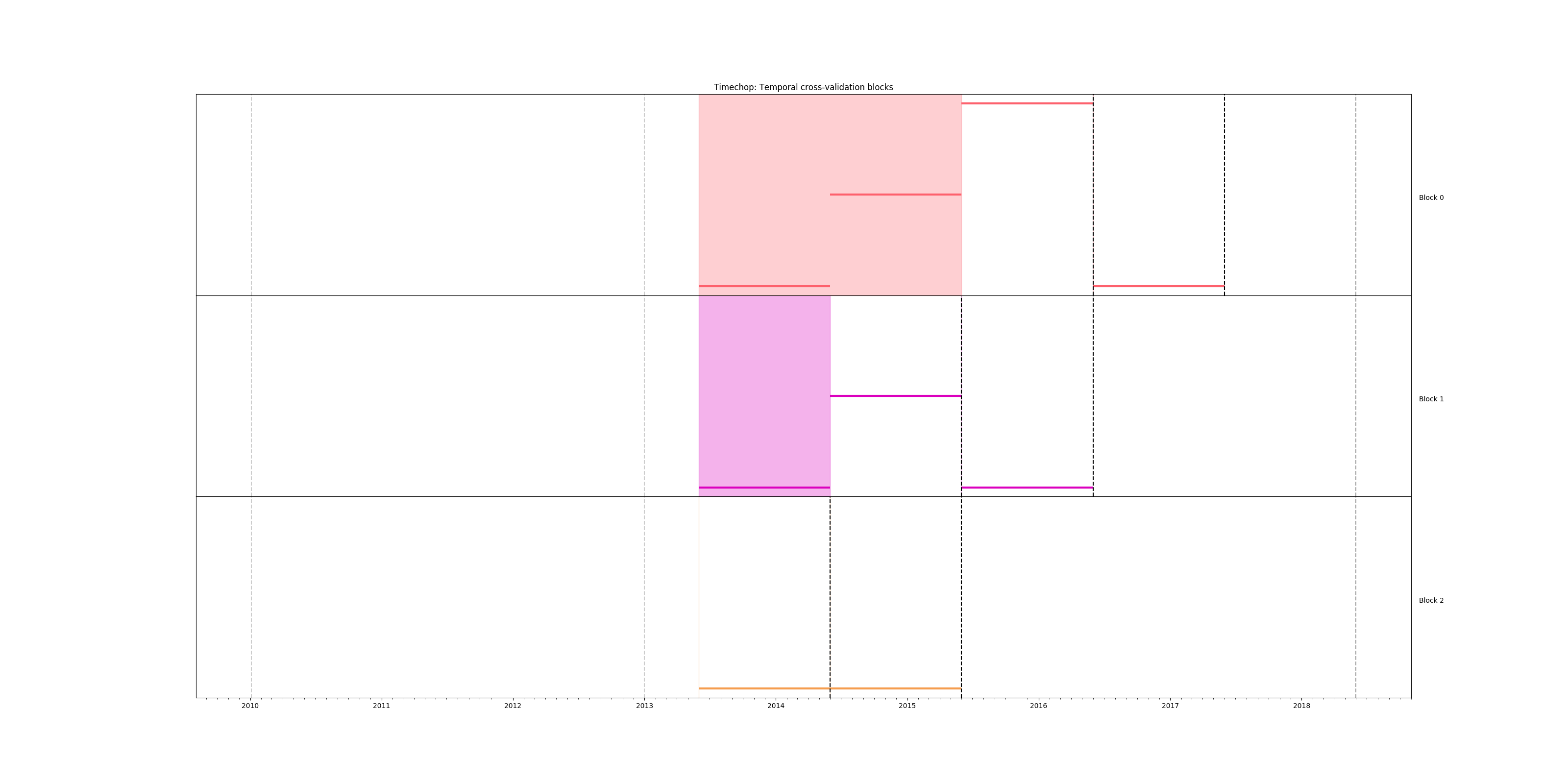 Figure. Temporal blocks
for inspections experiment. The label is a failed inspection in the
next month.
Figure. Temporal blocks
for inspections experiment. The label is a failed inspection in the
next month.
We want to use all the features groups
(feature_group_definition). The training will be made on matrices
with all the feature groups, then leaving one feature group out at a
time, leave-one-out (i.e. one model with inspections and
results, another with inspections and risks, and another with
results and risks), and finally leaving one feature group in at a
time (i.e. a model withinspectionsonly, another withresultsonly, and a third withrisks` only).
feature_group_definition:
prefix:
- 'inspections'
- 'results'
- 'risks'
- 'inspection_types'
feature_group_strategies: ['all', 'leave-one-in', 'leave-one-out']
Finally, we will try some RandomForestClassifier:
grid_config:
'sklearn.ensemble.RandomForestClassifier':
n_estimators: [10000]
criterion: ['gini']
max_depth: [2, 5, 10]
max_features: ['sqrt']
min_samples_split: [2, 10, 50]
n_jobs: [-1]
'sklearn.ensemble.ExtraTreesClassifier':
n_estimators: [10000]
criterion: ['gini']
max_depth: [2, 5, 10]
max_features: ['sqrt']
min_samples_split: [2, 10, 50]
n_jobs: [-1]
scoring:
testing_metric_groups:
-
metrics: [precision@, recall@]
thresholds:
percentiles: [1.0, 2.0, 3.0, 4.0, 5.0, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100]
top_n: [1, 5, 10, 25, 50, 100, 250, 500, 1000]
training_metric_groups:
-
metrics: [accuracy]
-
metrics: [precision@, recall@]
thresholds:
percentiles: [1.0, 2.0, 3.0, 4.0, 5.0, 10, 15, 20, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100]
top_n: [1, 5, 10, 25, 50, 100, 250, 500, 1000]
# Remember to run this in bastion NOT in your laptop shell!
triage experiment experiments/inspections_label_failed_01.yaml --validate-only
You can execute the experiment with
# Remember to run this in bastion NOT in your laptop shell!
time triage experiment experiments/inspections_label_failed_01.yaml
This will take a looooong time to run. The reason for that is easy to
understand: We are computing a lot of models: 6 time splits, 18 model
groups and 9 features sets (one for all, four for leave_one_in and
four for leave_one_out), so 6 \times 18 \times 9 = 486 extra
models.
Well, now we have a lot of models. How can you pick the best one? You could try the following query:
with features_groups as (
select
model_group_id,
split_part(unnest(feature_list), '_', 1) as feature_groups
from
triage_metadata.model_groups
),
features_arrays as (
select
model_group_id,
array_agg(distinct feature_groups) as feature_groups
from
features_groups
group by
model_group_id
)
select
model_group_id,
model_type,
hyperparameters,
feature_groups,
array_agg(to_char(stochastic_value, '0.999') order by train_end_time asc) filter (where metric = 'precision@') as "precision@15%",
array_agg(to_char(stochastic_value, '0.999') order by train_end_time asc) filter (where metric = 'recall@') as "recall@15%"
from
triage_metadata.models
join
features_arrays using(model_group_id)
join
test_results.evaluations using(model_id)
where
model_comment ~ 'inspection'
and
parameter = '15_pct'
group by
model_group_id,
model_type,
hyperparameters,
feature_groups
order by
model_group_id;
This is a long table …
| model_group_id | model_type | hyperparameters | feature_groups | precision@15% | recall@15% |
|---|---|---|---|---|---|
| 1 | sklearn.dummy.DummyClassifier | {"strategy": "prior"} | {inspections} | {" 0.339"," 0.366"," 0.378"} | {" 0.153"," 0.151"," 0.149"} |
| 2 | sklearn.tree.DecisionTreeClassifier | {"max_depth": 2, "min_samples_split": 2} | {inspection,inspections,results,risks} | {" 0.347"," 0.394"," 0.466"} | {" 0.155"," 0.153"," 0.180"} |
| 3 | sklearn.tree.DecisionTreeClassifier | {"max_depth": 2, "min_samples_split": 5} | {inspection,inspections,results,risks} | {" 0.349"," 0.397"," 0.468"} | {" 0.156"," 0.154"," 0.181"} |
| 4 | sklearn.tree.DecisionTreeClassifier | {"max_depth": 10, "min_samples_split": 2} | {inspection,inspections,results,risks} | {" 0.409"," 0.407"," 0.470"} | {" 0.179"," 0.163"," 0.178"} |
| 5 | sklearn.tree.DecisionTreeClassifier | {"max_depth": 10, "min_samples_split": 5} | {inspection,inspections,results,risks} | {" 0.416"," 0.409"," 0.454"} | {" 0.183"," 0.160"," 0.169"} |
| 6 | sklearn.tree.DecisionTreeClassifier | {"max_depth": null, "min_samples_split": 2} | {inspection,inspections,results,risks} | {" 0.368"," 0.394"," 0.413"} | {" 0.165"," 0.161"," 0.160"} |
| 7 | sklearn.tree.DecisionTreeClassifier | {"max_depth": null, "min_samples_split": 5} | {inspection,inspections,results,risks} | {" 0.386"," 0.397"," 0.417"} | {" 0.171"," 0.161"," 0.162"} |
| 8 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {inspection,inspections,results,risks} | {" 0.441"," 0.471"," 0.513"} | {" 0.190"," 0.187"," 0.193"} |
| 9 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {inspection,inspections,results,risks} | {" 0.470"," 0.478"," 0.532"} | {" 0.200"," 0.189"," 0.200"} |
| 10 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {inspection,inspections,results,risks} | {" 0.481"," 0.479"," 0.513"} | {" 0.204"," 0.189"," 0.193"} |
| 11 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {inspection,inspections,results,risks} | {" 0.474"," 0.472"," 0.535"} | {" 0.202"," 0.183"," 0.199"} |
| 12 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {inspections} | {" 0.428"," 0.417"," 0.389"} | {" 0.179"," 0.149"," 0.148"} |
| 13 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {inspections} | {" 0.428"," 0.417"," 0.390"} | {" 0.180"," 0.149"," 0.148"} |
| 14 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {inspections} | {" 0.427"," 0.417"," 0.376"} | {" 0.179"," 0.149"," 0.140"} |
| 15 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {inspections} | {" 0.428"," 0.417"," 0.380"} | {" 0.179"," 0.149"," 0.143"} |
| 16 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {results} | {" 0.415"," 0.398"," 0.407"} | {" 0.180"," 0.157"," 0.157"} |
| 17 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {results} | {" 0.393"," 0.401"," 0.404"} | {" 0.171"," 0.158"," 0.155"} |
| 18 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {results} | {" 0.436"," 0.425"," 0.447"} | {" 0.191"," 0.169"," 0.171"} |
| 19 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {results} | {" 0.432"," 0.423"," 0.438"} | {" 0.188"," 0.168"," 0.167"} |
| 20 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {risks} | {" 0.413"," 0.409"," 0.431"} | {" 0.184"," 0.170"," 0.166"} |
| 21 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {risks} | {" 0.407"," 0.391"," 0.459"} | {" 0.180"," 0.159"," 0.179"} |
| 22 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {risks} | {" 0.418"," 0.432"," 0.469"} | {" 0.184"," 0.176"," 0.181"} |
| 23 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {risks} | {" 0.427"," 0.431"," 0.476"} | {" 0.187"," 0.176"," 0.183"} |
| 24 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {inspection} | {" 0.435"," 0.483"," 0.483"} | {" 0.193"," 0.194"," 0.186"} |
| 25 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {inspection} | {" 0.448"," 0.465"," 0.518"} | {" 0.196"," 0.188"," 0.202"} |
| 26 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {inspection} | {" 0.446"," 0.446"," 0.508"} | {" 0.189"," 0.179"," 0.193"} |
| 27 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {inspection} | {" 0.459"," 0.444"," 0.513"} | {" 0.198"," 0.176"," 0.198"} |
| 28 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {inspection,results,risks} | {" 0.472"," 0.479"," 0.506"} | {" 0.202"," 0.191"," 0.190"} |
| 29 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {inspection,results,risks} | {" 0.476"," 0.486"," 0.532"} | {" 0.202"," 0.191"," 0.199"} |
| 30 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {inspection,results,risks} | {" 0.485"," 0.454"," 0.535"} | {" 0.203"," 0.180"," 0.204"} |
| 31 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {inspection,results,risks} | {" 0.479"," 0.497"," 0.521"} | {" 0.205"," 0.193"," 0.196"} |
| 32 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {inspection,inspections,risks} | {" 0.437"," 0.432"," 0.474"} | {" 0.191"," 0.178"," 0.181"} |
| 33 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {inspection,inspections,risks} | {" 0.459"," 0.468"," 0.501"} | {" 0.202"," 0.191"," 0.197"} |
| 34 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {inspection,inspections,risks} | {" 0.461"," 0.448"," 0.482"} | {" 0.201"," 0.181"," 0.187"} |
| 35 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {inspection,inspections,risks} | {" 0.463"," 0.445"," 0.497"} | {" 0.200"," 0.180"," 0.189"} |
| 36 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {inspection,inspections,results} | {" 0.462"," 0.448"," 0.513"} | {" 0.199"," 0.177"," 0.191"} |
| 37 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {inspection,inspections,results} | {" 0.465"," 0.491"," 0.537"} | {" 0.197"," 0.190"," 0.203"} |
| 38 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {inspection,inspections,results} | {" 0.459"," 0.481"," 0.522"} | {" 0.193"," 0.187"," 0.198"} |
| 39 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {inspection,inspections,results} | {" 0.474"," 0.479"," 0.536"} | {" 0.203"," 0.188"," 0.201"} |
| 40 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 2} | {inspections,results,risks} | {" 0.436"," 0.429"," 0.490"} | {" 0.189"," 0.174"," 0.185"} |
| 41 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 2} | {inspections,results,risks} | {" 0.441"," 0.448"," 0.515"} | {" 0.190"," 0.180"," 0.194"} |
| 42 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 100, "min_samples_split": 10} | {inspections,results,risks} | {" 0.460"," 0.475"," 0.481"} | {" 0.198"," 0.189"," 0.178"} |
| 43 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {inspections,results,risks} | {" 0.465"," 0.446"," 0.496"} | {" 0.199"," 0.179"," 0.187"} |
This table summarizes all our experiments, but it is very difficult to
use if you want to choose the best combination of hyperparameters and
algorithm (i.e. the model group). In the next section
will solve this dilemma with the support of audition.
Selecting the best model#
43 model groups! How to pick the best one and use it for making predictions with new data? What do you mean by “the best”? This is not as easy as it sounds, due to several factors:
- You can try to pick the best using a metric specified in the
config file (
precision@andrecall@), but at what point of time? Maybe different model groups are best at different prediction times. - You can just use the one that performs best on the last test set.
- You can value a model group that provides consistent results over time. It might not be the best on any test set, but you can feel more confident that it will continue to perform similarly.
- If there are several model groups that perform similarly and their lists are more or less similar, maybe it doesn't really matter which you pick.
Remember…
Before move on, remember the two main caveats for the value of the metric in this kind of ML problems:
- Could be many entities with the same predicted risk score (ties)
- Could be a lot of entities without a label (Weren't inspected, so we don’t know)
We included a simple configuration file in /triage/audition/inspection_audition_config.yaml
with some rules:
# CHOOSE MODEL GROUPS
model_groups:
query: |
select distinct(model_group_id)
from triage_metadata.model_groups
where model_config ->> 'experiment_type' ~ 'inspection'
# CHOOSE TIMESTAMPS/TRAIN END TIMES
time_stamps:
query: |
select distinct train_end_time
from triage_metadata.models
where model_group_id in ({})
and extract(day from train_end_time) in (1)
and train_end_time >= '2014-01-01'
# FILTER
filter:
metric: 'precision@' # metric of interest
parameter: '10_pct' # parameter of interest
max_from_best: 1.0 # The maximum value that the given metric can be worse than the best model for a given train end time.
threshold_value: 0.0 # The worst absolute value that the given metric should be.
distance_table: 'inspections_distance_table' # name of the distance table
models_table: 'models' # name of the models table
# RULES
rules:
-
shared_parameters:
-
metric: 'precision@'
parameter: '10_pct'
selection_rules:
-
name: 'best_current_value' # Pick the model group with the best current metric value
n: 3
-
name: 'best_average_value' # Pick the model with the highest average metric value
n: 3
-
name: 'lowest_metric_variance' # Pick the model with the lowest metric variance
n: 3
-
name: 'most_frequent_best_dist' # Pick the model that is most frequently within `dist_from_best_case`
dist_from_best_case: [0.05]
n: 3
Audition will have each rule give you the best n model groups
based on the metric and parameter following that rule for the most
recent time period (in all the rules shown n = 3).
We can run the simulation of the rules against the experiment as:
# Run this in bastion…
triage --tb audition -c inspection_audition_config.yaml --directory audition/inspections
Audition will create several plots that will help you to sort out
which is the best model group to use (like in a production setting
or just to generate your predictions list).
Filtering model groups#
Most of the time, audition should be used in a iterative fashion,
the result of each iteration will be a reduced set of models groups
and a best rule for selecting model groups.
If you look again at the audition configuration
file above
you can filter the number of models to consider using the parameters
max_from_best and threshold_value. The former will filter out
models groups with models which performance in the metric is
farther than the max_from_best (In this case we are allowing all
the models, since max_from_best = 1.0, if you want less models you
could choose 0.1 for example, and you will remove the
DummyClassifier and some
DecisionTreeClassifiers). threshold_value filter out all the
models groups with models performing below that the specified
value. This could be important if you don’t find acceptable models
with metrics that are that low.
Audition will generate two plots that are meant to be used together:
model performance over time and distance from best.
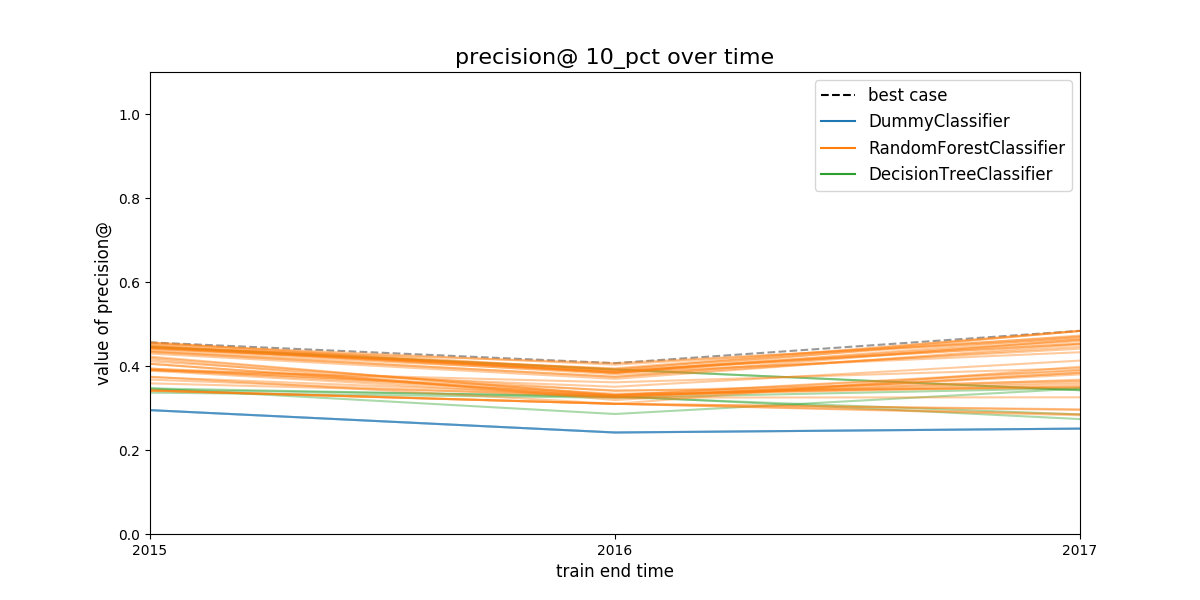 Figure. Model group performance over time. In this case the metric
show is
Figure. Model group performance over time. In this case the metric
show is precision@10%. We didn’t filter out any model group, so the
45 model groups are shown. See discussion above to learn how to plot
less model groups. The black dashed line represents the (theoretical)
system's performance if we select the best performant model in a every
evaluation date. The colored lines represents different model
groups. All the model groups that share an algorithm will be colored
the same.
Next figure shows the proportion of models that are behind the best
model. The distance is measured in percentual points. You could use
this plot to filter out more model groups, changing the value of
max_from_best in the configuration file. This plot is hard to read,
but is very helpful since it shows you the consistency of the model
group: How consistently are the model group in a specific range,
let's say 20 points, from the best?
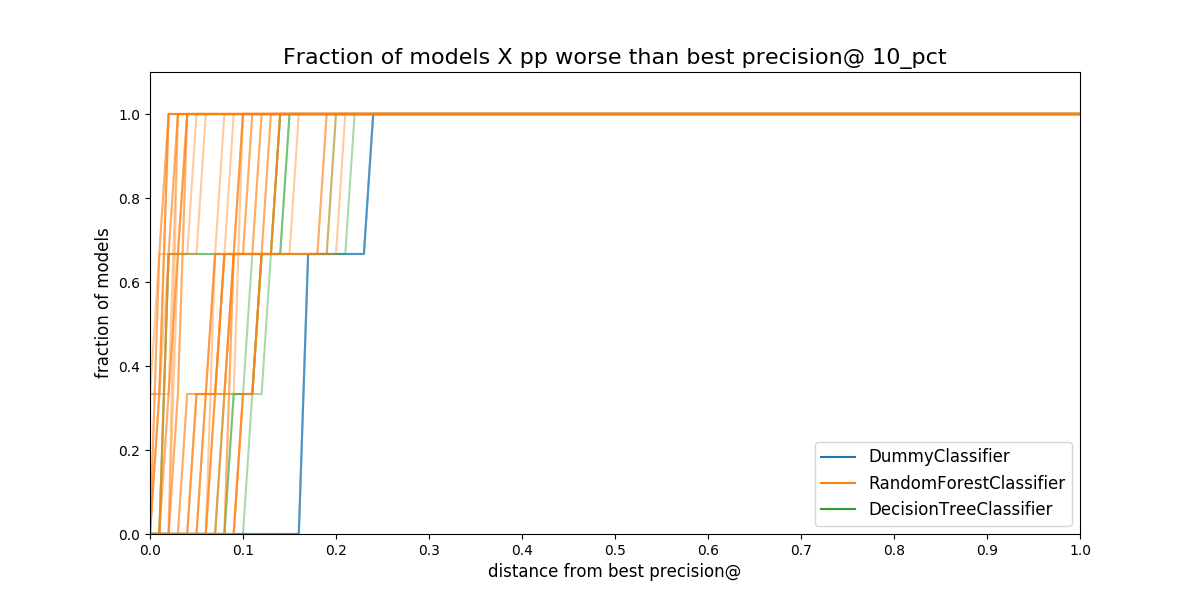 Figure. Proportion of *models in a model group that are separated
from the best model. The distance is measured in percentual points,
i.e. How much less precision at 10 percent of the population compared
to the best model in that date.*
Figure. Proportion of *models in a model group that are separated
from the best model. The distance is measured in percentual points,
i.e. How much less precision at 10 percent of the population compared
to the best model in that date.*
In the figure, you can see that the ~60% of the DummyClassifier
models are ~18 percentual points below of the best.
Selecting the best rule or strategy for choosing model groups#
In this phase of the audition, you will see what will happen in the next time if you choose your model group with an specific strategy or rule. We call this the regret of the strategies.
We define regret as
Regret Is the difference in performance between the model group you picked and the best one in the next time period.
The next plot show the best model group selected by the strategies specified in the configuration file:
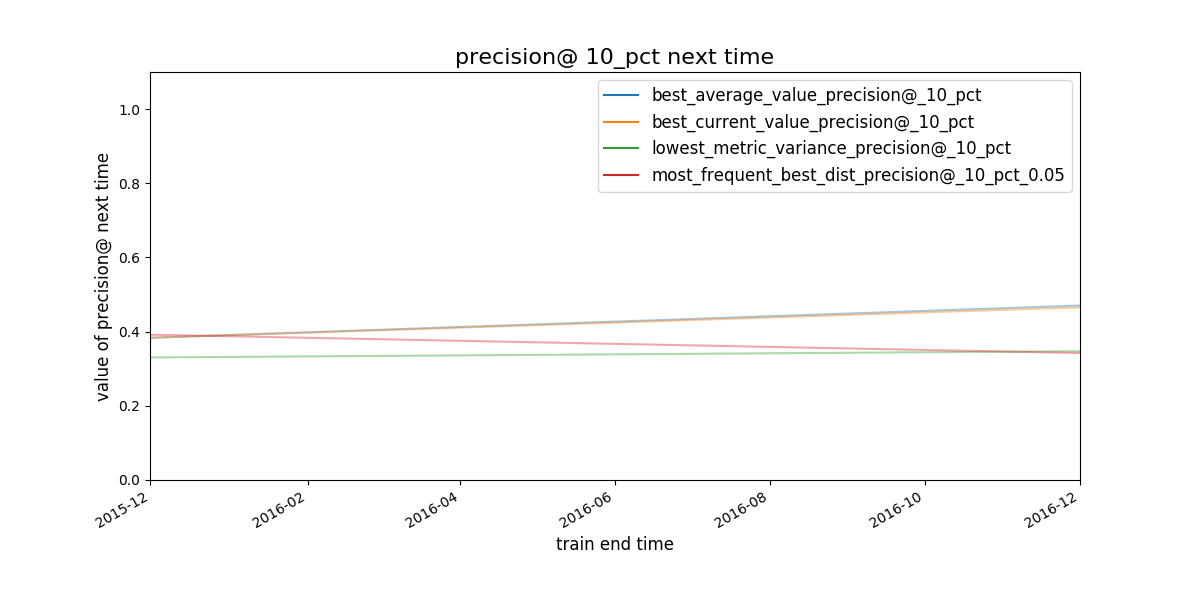 Figure. Given a strategy for selecting model groups (in the figure, 4
are shown), What will be the performace of the model group chosen by
that strategy in the next evaluation date?
Figure. Given a strategy for selecting model groups (in the figure, 4
are shown), What will be the performace of the model group chosen by
that strategy in the next evaluation date?
It seems that the strategies best average and best current value select the same model group.
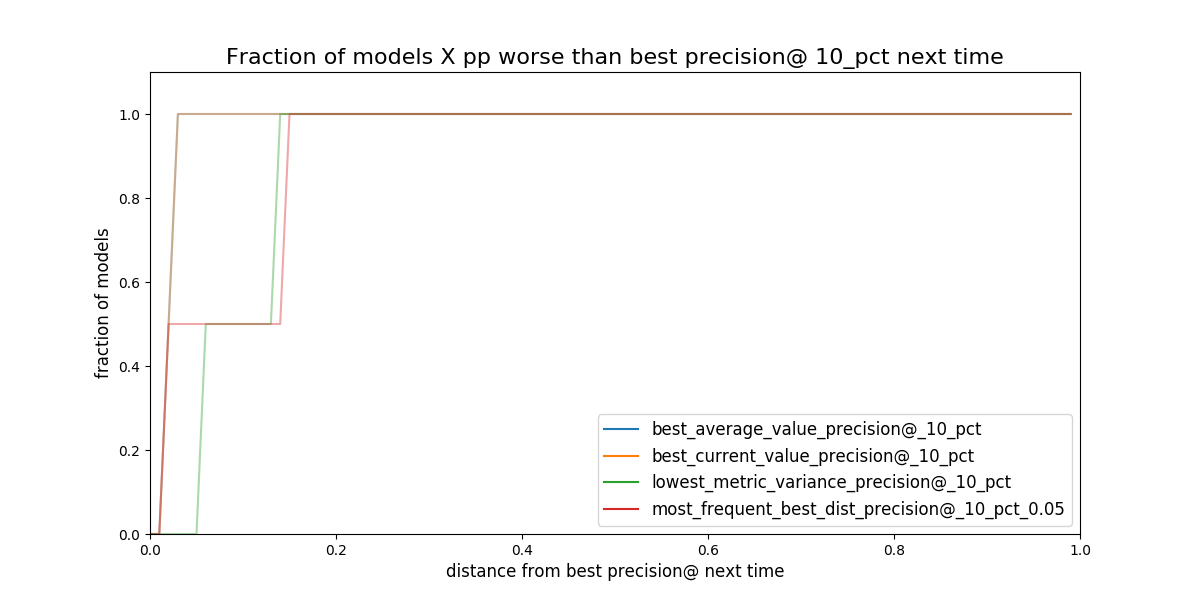 Figure. Given a strategy for selecting model groups (in the plot 4
are shown). What will be the distance (*regret) to the best
theoretical model in the following evaluation date?*
Figure. Given a strategy for selecting model groups (in the plot 4
are shown). What will be the distance (*regret) to the best
theoretical model in the following evaluation date?*
Obviously, you don’t know the future, but with the available data, if you stick to an a particular strategy, How much you will regret about that decision?
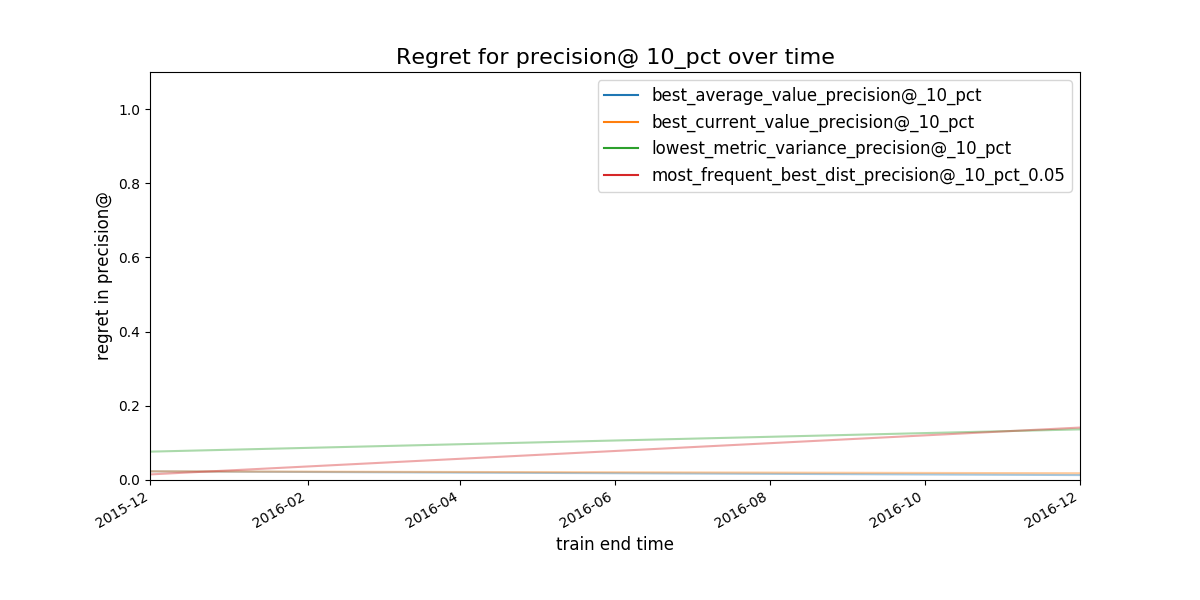 Figure. Expected regret for the strategies. The less the better.
Figure. Expected regret for the strategies. The less the better.
The best 3 model groups per strategy will be stored in the file [[file:audition/inspections/results_model_group_ids.json][results_model_group_ids.json]]:
{
"best_current_value_precision@_10_pct": [39, 30, 9],
"best_average_value_precision@_10_pct": [39, 9, 29],
"lowest_metric_variance_precision@_10_pct": [1, 5, 19],
"most_frequent_best_dist_precision@_10_pct_0.05": [8, 9, 10]
}
The analysis suggests that the best strategies are
- select the model groups (
39,9,29) which have the best average precision@10% value or, - select the best model group (
39,30,9) using precision@10% today and use it for the next time period.
You will note that both strategies share two models groups and differ
in one. In the next two sections, we will investigate further those
four model groups selected by audition, using the Postmodeling
tool set.
Postmodeling: Inspecting the best models closely#
Postmodeling will help you to understand the behaviour orf your selected models (from audition)
As in Audition, we will split the postmodeling process in two
parts. The first one is about exploring the model groups filtered by
audition, with the objective of select one. The second part is about
learning about models in the model group that was selected.
We will setup some parameters in the postmodeling configuration
file located at /triage/postmodeling/inspection_postmodeling_config.yaml,
mainly where is the audition’s output file located.
# Postmodeling Configuration File
project_path: '/triage' # Project path defined in triage with matrices and models
model_group_id:
- 39
- 9
- 29
- 30
thresholds: # Thresholds for defining positive predictions
rank_abs: [50, 100, 250]
rank_pct: [5, 10, 25]
baseline_query: | # SQL query for defining a baseline for comparison in plots. It needs a metric and parameter
select g.model_group_id,
m.model_id,
extract('year' from m.evaluation_end_time) as as_of_date_year,
m.metric,
m.parameter,
m.value,
m.num_labeled_examples,
m.num_labeled_above_threshold,
m.num_positive_labels
from test_results.evaluations m
left join triage_metadata.models g
using(model_id)
where g.model_group_id = 1
and metric = 'precision@'
and parameter = '10_pct'
max_depth_error_tree: 5 # For error trees, how depth the decision trees should go?
n_features_plots: 10 # Number of features for importances
figsize: [12, 12] # Default size for plots
fontsize: 20 # Default fontsize for plots
Compared to the previous sections, postmodeling is not an automated process (yet). Hence, to do the following part of the tutorial, you need to run jupyter inside bastion as follows:
jupyter-notebook –-ip=0.0.0.0 --port=56406 --allow-root
And then in your browser type11: http://0.0.0.0:56406
Now that you are in a jupyter notebook, type the following:
%matplotlib inline
import matplotlib
#matplotlib.use('Agg')
import triage
import pandas as pd
import numpy as np
from collections import OrderedDict
from triage.component.postmodeling.contrast.utils.aux_funcs import create_pgconn, get_models_ids
from triage.component.catwalk.storage import ProjectStorage, ModelStorageEngine, MatrixStorageEngine
from triage.component.postmodeling.contrast.parameters import PostmodelParameters
from triage.component.postmodeling.contrast.model_evaluator import ModelEvaluator
from triage.component.postmodeling.contrast. model_group_evaluator import ModelGroupEvaluator
After importing, we need to create an sqlalchemy engine for connecting to the database, and read the configuration file.
params = PostmodelParameters('inspection_postmodeling_config.yaml')
engine = create_pgconn('database.yaml')
Postmodeling provides the object ModelGroupEvaluator to compare different model groups.
audited_models_class = ModelGroupEvaluator(tuple(params.model_group_id), engine)
Comparing the audited model groups#
First we will compare the performance of the audited model groups and the baseline over time. First, we will plot precision@10_pct
audited_models_class.plot_prec_across_time(param_type='rank_pct',
param=10,
baseline=True,
baseline_query=params.baseline_query,
metric='precision@',
figsize=params.figsize)
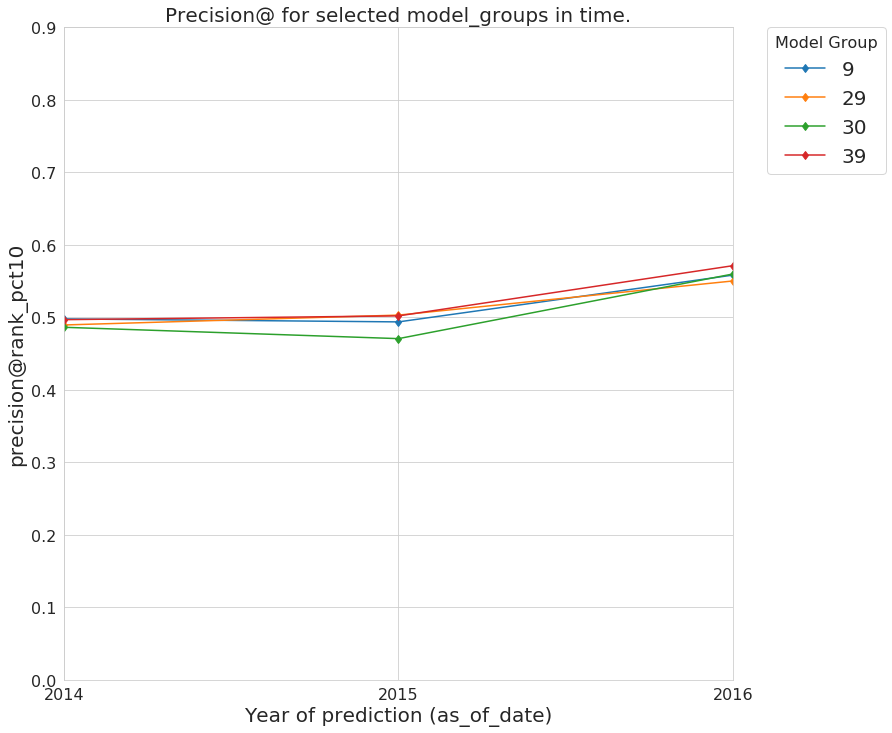 Figure. Precision@10% over time from the best performing model groups selected by Audition
Figure. Precision@10% over time from the best performing model groups selected by Audition
and now the recall@10_pct
audited_models_class.plot_prec_across_time(param_type='rank_pct',
param=10,
metric='recall@',
figsize=params.figsize)
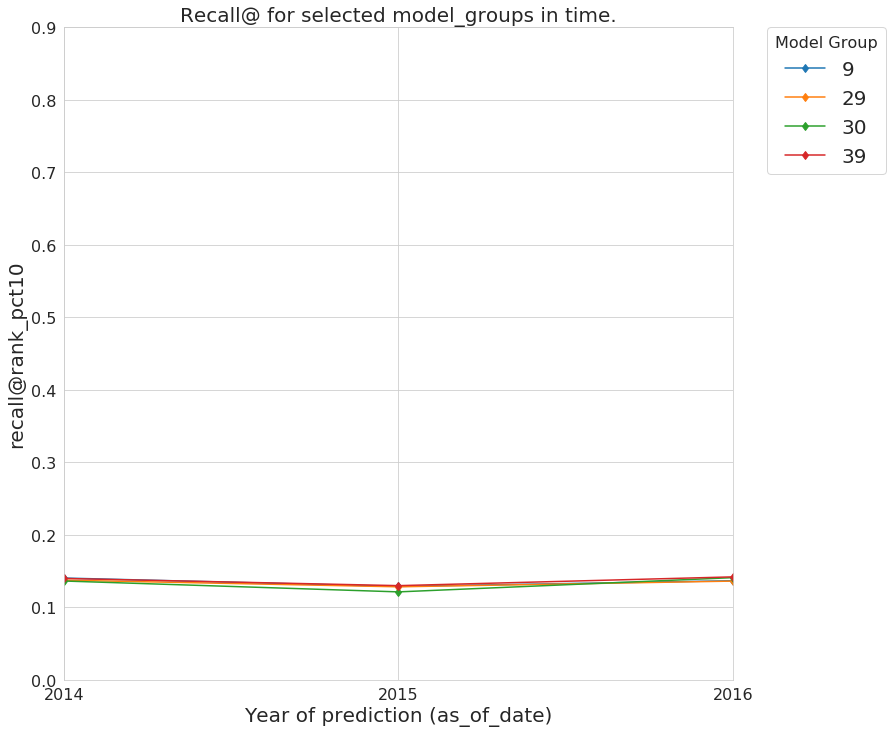 Figure. Recall@10% over time from the best performing model groups selected by Audition
Figure. Recall@10% over time from the best performing model groups selected by Audition
All the selected model groups have a very similar performance. Let’s see if they are predicting similar lists of facilities that are at risk of fail an inspection.
audited_models_class.plot_jaccard_preds(param_type='rank_pct',
param=10,
temporal_comparison=True)
 Figure. How similar are the model groups’ generated list? We use
Jaccard similarity on the predicted lists (length of list 10%) to
asses the overlap between lists.
Figure. How similar are the model groups’ generated list? We use
Jaccard similarity on the predicted lists (length of list 10%) to
asses the overlap between lists.
The plot will shows the overlap of the predicted list containing the 10% of the facilities between model groups at each as of date. The lists are at least 50% similar.
Tip
Why the models are not learning the same? You should investigate why is that so. This could lead you to defining new features or some another conclusion about your data, but for this tutorial we will move on.
Going deeper with a model#
Imagine that after a deeper analysis, you decide to choose model group 39
select
mg.model_group_id,
mg.model_type,
mg.hyperparameters,
array_agg(model_id order by train_end_time) as models
from
triage_metadata.model_groups as mg
inner join
triage_metadata.models
using (model_group_id)
where model_group_id = 39
group by 1,2,3
| model_group_id | model_type | hyperparameters | models |
|---|---|---|---|
| 39 | sklearn.ensemble.RandomForestClassifier | {"criterion": "gini", "max_features": "sqrt", "n_estimators": 250, "min_samples_split": 10} | {53,89,125} |
We will investigate what the particular models are doing. Postmodeling created a ModelEvaluator (similar to the ModelGroupEvaluator) to do this exploration:
models_39 = { f'{model}': ModelEvaluator(39, model, engine) for model in [53,89,125] }
In this tutorial, we will just show some parts of the analysis in the most recent model, but feel free of exploring the behavior of all the models in this model group, and check if you can detect any pattern.
-
Feature importances
models_39['125'].plot_feature_importances(path=params.project_path, n_features_plots=params.n_features_plots, figsize=params.figsize)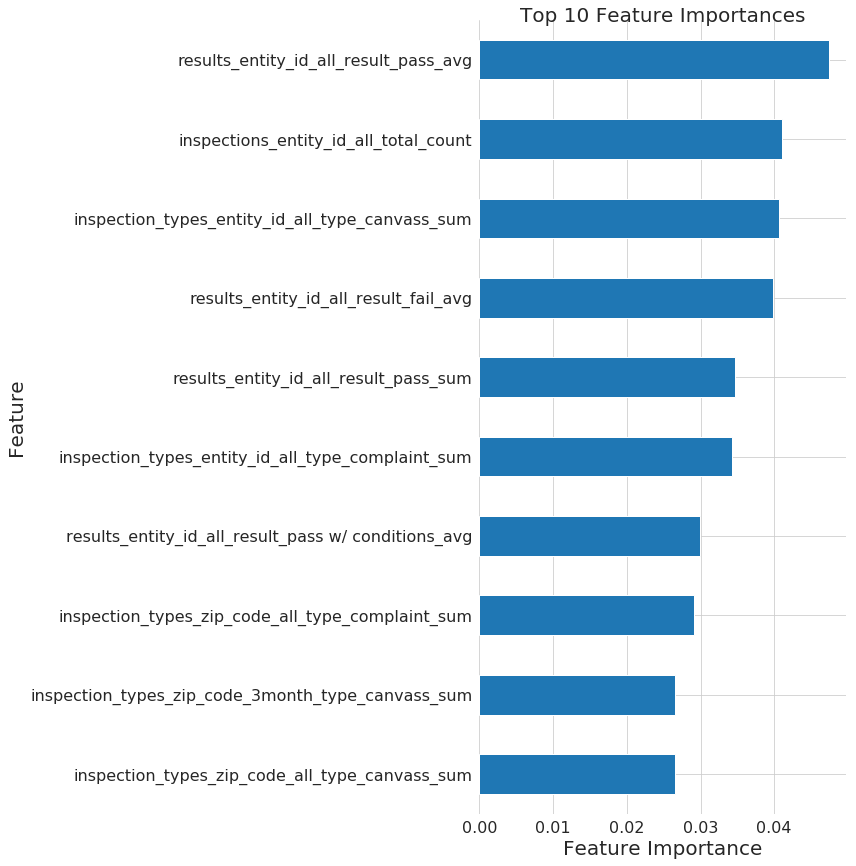 Figure. Top 10 feature importances for de model group 11 at 2016-06-01 (i.e. model 125).
Figure. Top 10 feature importances for de model group 11 at 2016-06-01 (i.e. model 125).models_39['125'].plot_feature_group_aggregate_importances()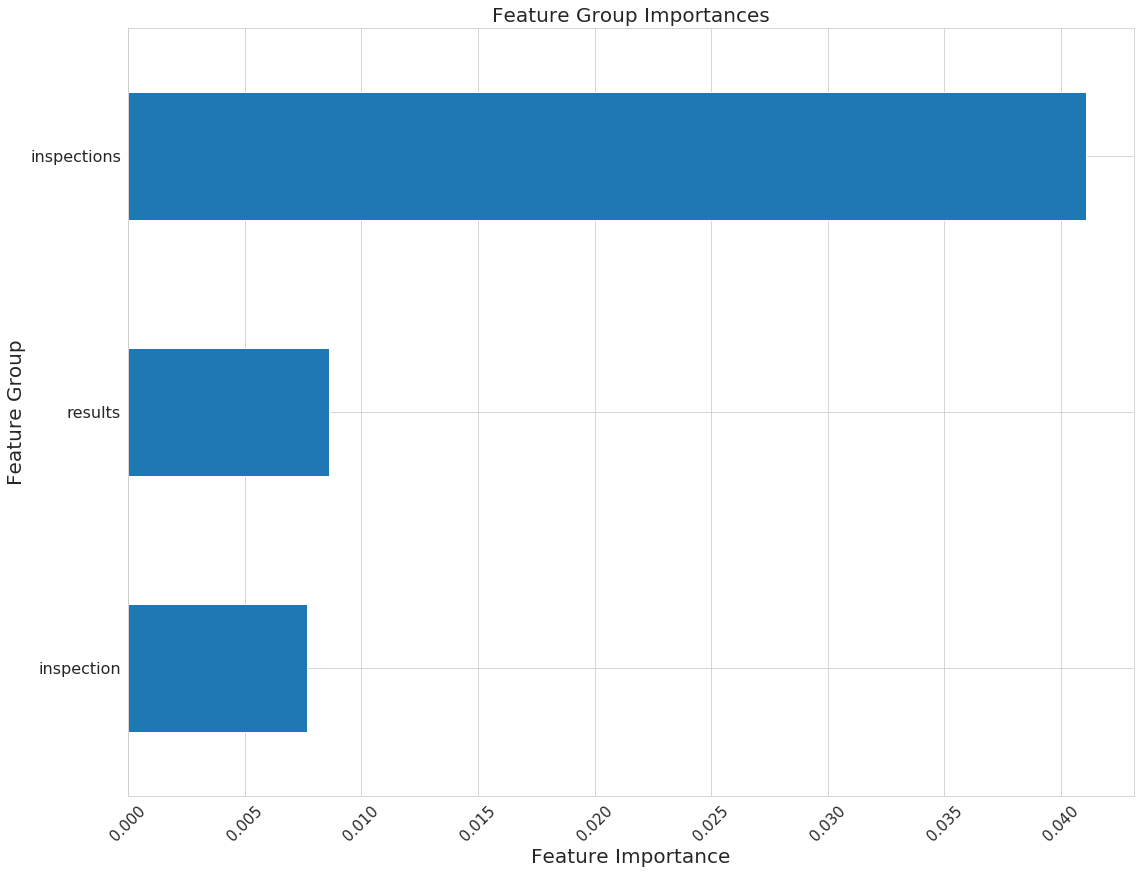 Figure. Feature group “importance” (we are basically taking the
max of all the feature importances in a feature group) for the
model group 39, model 125.
Figure. Feature group “importance” (we are basically taking the
max of all the feature importances in a feature group) for the
model group 39, model 125. -
Our Policy menu
The following plot depicts the behavior of the metrics if you change the length of the facilities predicted at risk (i.e. the k). This plot is important from the decision making point of view, since it could be used as a policy menu.
models_39['125'].plot_precision_recall_n()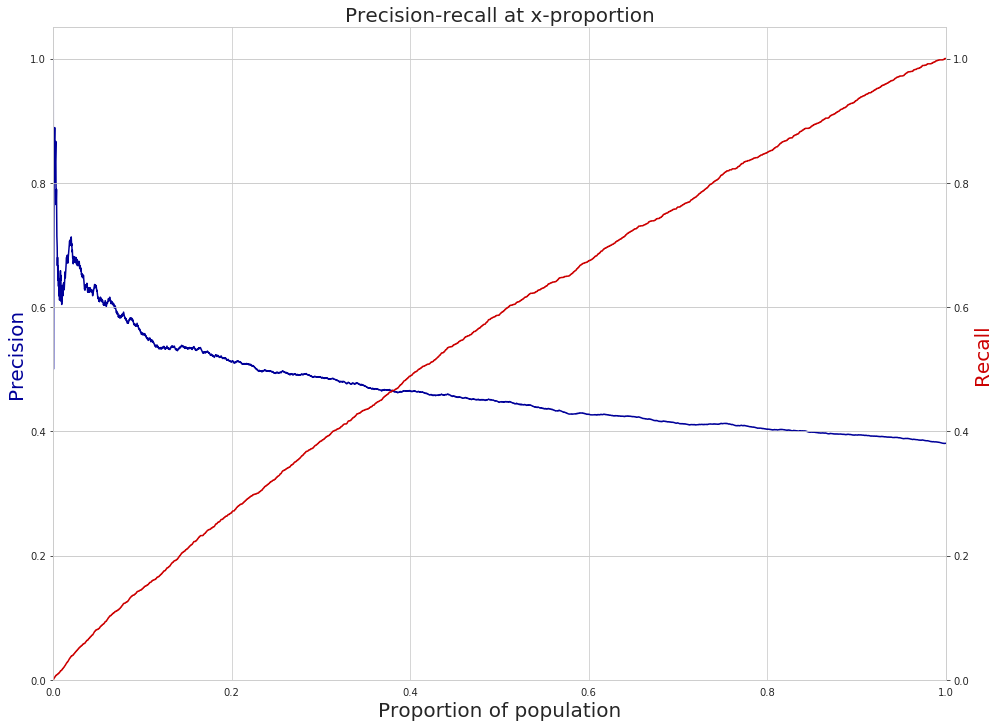 Figure. Plot of Precision and Recall over the proportion of the
facilities. This plot is used as a "policy menu" since allows you
to see how much you will gain if you invest more resources or how
much you will sacrifice if you reduce your budget for
resources. This is also known as “Rayid plot” at DSaPP.
Figure. Plot of Precision and Recall over the proportion of the
facilities. This plot is used as a "policy menu" since allows you
to see how much you will gain if you invest more resources or how
much you will sacrifice if you reduce your budget for
resources. This is also known as “Rayid plot” at DSaPP.We selected this model group because it was the best at precision at 10% (i.e. the model group consistently chose facilities will fail inspections at the top 10% of the risk). With the plot above you could decide to double your resources (maybe hiring more inspectors so you could inspect 20% of the facilities) and with this model you will double the detection of facilities that will fail inspections (from ~18% to ~30% in recall) with only a few percent points less of precision ~45% to ~40% (this means that 6 in 10 facilities that the inspectors visit will pass the inspection). You could also go the other way around: if you reduce the length of the list from 10% to 5%, well you will gain a little of precision, but your recall will be ~5%.
Where to go from here#
Ready to get started with your own data? Check out the suggested project workflow for some tips about how to iterate and tune the pipeline for your project.
Want to work through another example? Take a look at our early warning system case study
-
If you assume a uniform distribution, it will make sense to select facilities at random. ↩
-
The underlying assumption here is that the City of Chicago is currently doing random selection for the inspections. This is not true (and probably unfair). In a real project, you will setup a real baseline and you will compare your models against it. This baseline could be a rule or a model. ↩
-
You need to check this! Fortunately,
triageallows you to try several options here, so, if you think that this is too high or too low you can change that and fit your needs. ↩ -
Think about it: we can’t learn the relationship between the features and the label if we don't know the label. ↩
-
Confused? Check A deeper look at triage for more details. ↩
-
The formulas are, for
precision@k, is the proportion of facilities correctly identified in the top-k facilities ranked by risk:$$ precision@k = \frac{TP \in k}{k} $$
This is a measure about how efficiently are your system using your resources.
recall@k, in the other hand is the proportion of all the facilities that are risk found in the top-k$$ recall@k = \frac{TP \in k}{TP} $$
recall is a measure about the coverage of your system, i.e. how good is identifying in the top-k the facilities at risk.
One possible variation of this is to only include in the denominator the labeled rows in k. This is the approach used by
triage. ↩ -
We will explore how to one way to tackle this in the advance part of this tutorial. ↩
-
The flags
-no-save-predictionsandprofileare not necessary but useful. The first one configure triage to not store the predictions (at this stage you don't need them, and you can always could recreate them from the model and the matrix). This will save you execution time. The flagprofilestores the execution profile times in a file, so you can check which models or matrices are taking a lot of time on been built. ↩ -
From a more mathematical point of view: Your data actually reflects the empirical probability: P(violation|inspected), i.e. the probability of find a violation given that the facility is inspected. But the probability that you want is P(violation) (yes, I know that there are no such things as unconditional probabilities, please bare with me),i.e. the probability that the facility is in violation. ↩
-
You should see that this assumption is very dangerous in other settings, for example, crime prediction. ↩
-
This assumes that you are in a GNU/Linux machine, if not (you should reconsider what are you doing with your life) you should change the ip address (
0.0.0.0) and use the one from the docker virtual machine. ↩ -
For some reason,
sklearndoesn’t scale the inputs to the Logistic Regression, so we (DSaPP) developed a version that does that. ↩